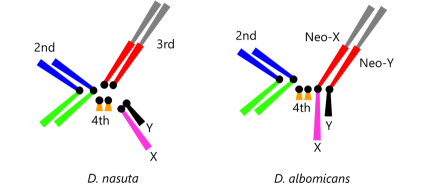
Evolutionary changes of genomic DNA sequences are an ultimate driving force of organismal evolutions. Therefore, it is important to study how DNA sequences change during the evolutionary period for a deep understanding of the evolution of genomes and organisms as well as performing accurate inferring phylogenetic trees and estimating divergence times. We are studying the molecular evolution of genes and genomes, using mathematical modeling, computer simulations, and real data analyses. A special focus is on developing methods for molecular evolutionary and phylogenetic analyses. The current project is to estimate divergence times in a phylogenetic tree in case of unequal evolutionary rates among lineages. [
Ref. 3][
Ref. 4]
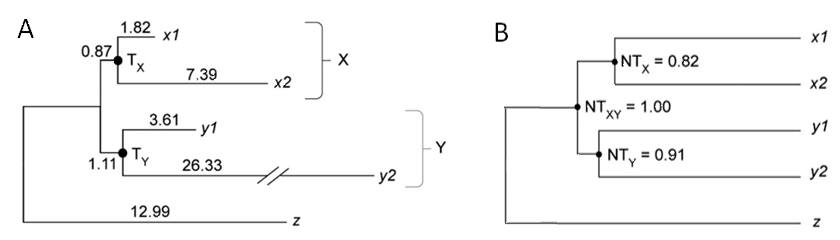
The phylogenetic tree of zinc finger protein genes from human and mouse, showing extreme evolutionary rate heterogeneity among genes and lineages (A), is successfully time-aligned by the RelTime method (B). x1: human ZFX, x2: mouse ZFX, y1: human ZFY, y2: mouse ZFY.