|
|
 Evolutionary Genetics Laboratory
Evolutionary Genetics Laboratory
|
Our research interests are the genetic basis of biological evolution and speciation. We use a variety of approaches ranging from field work to bioinformatic analyses as well as laboratory experiments.
|
|
|
Koichiro Tamura |
e-mail |
|
|
|
Aya Takahashi |
e-mail |
|
|
|
Masafumi Nozawa |
e-mail |
|
|
|
Molecular basis of adaptive evolution in Drosophila
|
|
Adaptation to temperate climate by a tropical species, D. albomicans The distribution of D. albomicans was limited in tropics in Southeast Asia until the mid-1980. Since then, however, the distribution has been extended toward north to west Japan to date. In our study, we found that the cold tolerance of this species has been improved in Japanese population with higher response to cold acclimation. This suggests that the improved cold tolerance is attributable to gene expression changes in response to the cold acclimation. Using RNA-seq method, we found many candidate genes responsible for the improvement of cold tolerance. We are trying to identify the causative genes among these genes by artificially modifying gene expression using GAL4/UAS system in D. melanogaster to clarify the molecular mechanism of cold acclimation response to enhance cold tolerance. Visit Dr. Tamura's web page for more info.
|
|
|
Molecular Evolution of DNA Sequences and Bioinformatics
|
|
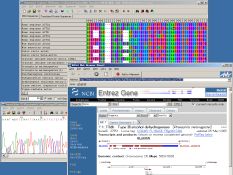
Bioinformatics for DNA sequence data
Genome sequencing projects have produced large amounts of DNA sequence data. Analyzing these data together with our original sequence data by computational and bioinformatic techniques, we are studying how DNA sequences, genes and genomes evolve. We are also developing new theoretical methods and computer software for molecular evolutionary and phylogenetic analyses. Visit Dr. Tamura's web page for more info.
|
|
|
Molecular bases of speciation in Drosophila
|
|
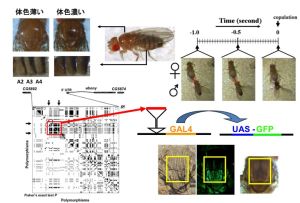
Experimental approaches using fruitflies
Studying the underlying molecular bases of speciation is one of the fundamental theme in evolutionary biology. Our aim is to understand the genetic changes that occur during the speciation event in Drosophila species. One of our current projects focuses on association between pigmentation intensity and sexual behavior in individuals from natural populations of D. melanogaster. We are also interested in analyzing the genomic features of transcriptome that could potentially influence the species isolation process. We take both experimental and bioinformatics approaches to tackle these questions.
|
|
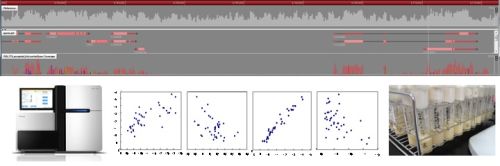
Genome-wide transciptome analyses using next generation sequencers
|
|
|
|
Evolutionary process of sex chromosomes in Drosophila
|
|
Sex chromosomes have repeatedly and independently emerged in many organisms during long history of life. After the emergence of sex chromosomes, however, Y chromosomes (or W chromosomes in the ZW sex-determining system) are degenerated in many cases, which would potentially be deleterious to males (or females in the ZW system). Then, how organisms have acquired and maintained the sex chromosomes under such a potential disadvantage? In our laboratory, we are tackling this issue using Drosophila miranda, D. albomicans, and D. americana, who recently acquired sex chromosomes. These young sex chromosomes are expected to be in the transient stages from ordinary autosomes to sex chromosomes, which is an ideal system to clarify the detailed processes of becoming sex chromosomes. We also study the evolutionary mechanisms of miRNA-involved gene regulatory networks. Next Generation Sequencing (or NGS, e.g., DNA-seq, RNA-seq, ChIP-seq, smallRNA-seq, CLIP-seq, etc.) and bioinformatics are keywords for both projects. Click here for more detailed information.
|
|
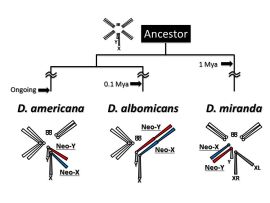
Drosophila species with young sex chromosomes
|
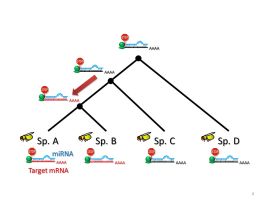
An empirical approach of studying evolution of miRNA-target pairs
|
|
|
|
Recent Publications
|
|
- Massey JH, Akiyam N, Bien T, Dreisewerd K, Wittkopp PJ, Yew JY, Takahashi A (2019) Pleiotropic effects of ebony and tan on pigmentation and cuticular hydrocarbon composition in Drosophila melanogaster. bioRxiv 538090.
- Moriguchi N, Uchiyama K, Miyagi R, Moritsuka E, Takahashi A, Tamura K, Tsumura Y, Teshima KM, Tachida H, Kusumi J (2019) Inferring the demographic history of Japanese cedar, Cryptomeria japonica, using amplicon sequencing. Heredity 123:371–383.
- Tao Q, Tamura K, Battistuzzi FU, Kumar S (2019) A machine learning method for detecting autocorrelation of evolutionary rates in large phylogenies. Mol. Biol. Evol. 36:811–824.
- Battistuzzi FU, Tao Q, Jones L, Tamura K, Kumar S (2018) RelTime relaxes the strict molecular clock throughout the phylogeny. Genome Biol. Evol. 10:1631-1636 2.
- Patel R, Scheinfeldt LB, Sanderford MD, Lanham TR, Tamura K, Platt A, Glicksberg BS, Xu K, Dudley JT, Kumar S (2018) Adaptive landscape of protein variation in human exomes. Mol. Biol. Evol. 35:2015-2025.
- Kumar S, Stecher G, Li M, Knyaz C, Tamura K (2018) MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 35:1547-1549.
- Tamura K, Tao Q, Kumar S (2018) Theoretical foundation of the RelTime method for estimating divergence times from variable evolutionary rates. Mol. Biol. Evol. 35:1770-1782
- Muto L, Kamimura Y, Tanaka KM, Takahashi A (2018) An innovative ovipositor for niche exploitation impacts genital coevolution between sexes in a fruit-damaging Drosophila. Proc. Royal Soc. B. 285:20181635
- Tanaka KM, Kamimura Y, Takahashi A (2018) Mechanical incompatibility caused by modifications of multiple male genital structures using genomic introgression in Drosophila. Evolution 72 (11), 2406-2418.
- Yuyama I, Ishikawa M, Nozawa M, Yoshida MA, Ikeo K (2018) Transcriptomic changes with increasing algal symbiont reveal the detailed process underlying establishment of coral-algal symbiosis. Sci. Rep. 8:16802.
- Matsunami M, Nozawa M, Suzuki R, Toga K, Masuoka Y, Yamaguchi K, Maekawa K, Shigenobu S, Miura T (2018) Caste-specific microRNA expression in termites: insights into soldier differentiation. Insect Mol. Biol. 28:86-98.
- Nozawa M, Ikeo K, Gojobori T (2018) Gene-by-gene or localized dosage compensation on the neo-X chromosome in Drosophila miranda. Genome Biol. Evol. 10:1875-1881.
- Sultana Z, Asakura A, Kinjo S, Nozawa M, Nakano T, Ikeo K (2018) Molecular phylogeny of ten intertidal hermit crabs of the genus Pagurus inferred from multiple mitochondrial genes, with special emphasis on evolutionary relationship of Pagurus lanuginosus and Pagurus maculosus. Genetica 146:369-381.
- Yamaguchi R, Yamanaka T, Liebhold AM (2019) Consequences of hybridization during the invasion on establishment success. Theor. Ecol. 1-9.
- Yamaguchi R, Suefuji S, Odagiri K, Peggie D, Yata O (2018) A color pattern difference in the fifth instar larva of two subspecies of Faunis menado Hewitson (Lepidoptera, Nymphalidae). Lepido. Sci. 69:67-73.
- Booton RD, Yamaguchi R, Iwasa Y (2018) A population model for diapausing multivoltine insects under asymmetric cannibalism. Popul. Ecol. 61:35-44.
- Booton R D, Yamaguchi R, Marshall J A R, Childs D Z, Iwasa Y (2018) Interactions between immunotoxicants and parasite stress: implications for host health. J. Theor. Biol. 445:120-127.
- Shimmori Y, Kanesaki Y, Nozawa M, Yoshikawa H, Ehira S*. (2018) Transcriptional activation of glycogen catabolism and oxidative pentose phosphate pathway by NrrA facilitates cell survival under nitrogen starvation in the cyanobacterium Synechococcus sp. strain PCC 7002. Plant Cell Physiol. 59:1225-1233.
- Hanada K*, Tezuka A, Nozawa M, Suzuki Y, Sugano S, Nagano AJ, Ito M, Morinaga S-I. (2018) Functional divergence of duplicate genes several million years after gene duplication in Arabidopsis. DNA Res. 25:327-339.
- Noyszewski AK,Liu YC,Tamura K,Smith AG. (2017) Polymorphism and structure of style–specific arabinogalactan proteins as determinants of pollen tube growth in Nicotiana. BMC evolutionary biology 17:186.
- Loh SYM,Ogawa Y,Kawana S,Tamura K,Lee HK. (2017) Semi-automated quantitative Drosophila wings measurements. BMC bioinformatics 18:319.
- Osada N, Miyagi R, Takahashi A* (2017) Cis- and trans-regulatory effects on gene expression in a natural population of Drosophila melanogaster. Genetics 206: 2139-2148.
- Takada Y, Miyagi R, Takahashi A, Endo T, Osada N* (2017) A generalized linear model for decomposing cis-regulatory, parent-of-origin, and maternal effects on allele-specific gene expression. G3 7: 2227-2234.
- Fraimout A, Debat V, Fellous S, Hufbauer RA, Foucaud J, Pudlo P, Marin J-M, Price DK, Cattel J, Chen X, Deprá M, Duyck PF, Guedot C, Kenis M, Kimura MT, Loeb G, Loiseau A, Martinez-Sañudo I, Pascual M, Richmond MP, Shearer P, Singh N, Tamura K, Xuéreb A, Zhang J, Estoup A. (2017) Deciphering the routes of invasion of Drosophila suzukii by means of ABC random forest. Mol. Biol. Evol. 34:980-996.
- Igawa T, Nozawa M, Suzuki DG, Reimer JD, Norov AR, Wang Y, Henmi Y, Yasui K*. (2017) Evolutionary history of the extant amphioxus lineage with shallow-branching diversification. Sci. Rep. 7:1157.
- Mello B, Tao Q, Tamura K, Kumar S. (2017) Fast and accurate estimates of divergence times from big data. Mol. Biol. Evol. 34:45-50.
- Fukushima K, Fang X, Alvarez-Ponce D, Cai H, Carretero-Paulet L, Chen C, Chang T-H, Farr KM, Fujita T, Hiwatashi Y, Hoshi Y, Imai T, Kasahara M, Librado P, Mao L, Mori H, Nishiyama T, Nozawa M, Palfalvi G, Pollard ST, Rozas J, Sanchez-Gracia A, Sankoff D, Shibata TF, Shigenobu S, Sumikawa N, Uzawa T, Xie M, Zheng C, Pollock DD, Albert VA, Li S, Hasebe M*. (2017) Genome of the pitcher plant Cephalotus reveals genetic changes associated with carnivory. Nature Ecol. Evol. 1:0059.
- Kudo A, Shigenobu S, Kadota K, Nozawa M, Shibata FT, Ishikawa Y, Matsuo T*. (2017) Comparative analysis of the brain transcriptome in a hyper-aggressive fruit fly, Drosophila prolongata. Insect Biochem. Mol. Biol. 82:11-20.
- Okamiya H*, Igawa T, Nozawa M, Sumida M, Kusano T. (2017) Development and Characterization of 23 Microsatellite Markers for the Montane Brown Frog (Rana ornativentris) Curr. Herpetol. 36:63-68.
- Kumar S, Stecher, G, Tamura K. (2016) MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol. Biol. Evol. 33:1870-1874.
- Sunaga S, Akiyama N, Miyagi R and Takahashi A*. (2016) Factors underlying natural variation in body pigmentation of Drosophila melanogaster. Genes Genet. Syst. 91: 127-137.
- Yoshida K, Miyagi R, Mori S, Takahashi A, Makino T, Toyoda A, Fujiyama A and Kitano J* (2016) Whole genome sequencing reveals small genomic regions of introgression in an introduced crater lake population of threespine stickleback. Ecol. Evol. 6: 2190-2204.
- Nozawa M*, Onizuka K, Fujimi M, Ikeo K, Gojobori T. (2016) Accelerated pseudogenization on the neo-X chromosome in Drosophila miranda. Nat. Commun. 7:13659.
- Komaki S, Lin S-M, Nozawa M, Oumi S, Sumida M, Igawa T*. (2016) Fine-scale demographic processes resulting from multiple overseas colonization events of the Japanese stream tree frog, Buergeria japonica. J Biogeogr. 12922.
- Ishikawa M, Shimizu H, Nozawa M, Ikeo K, Gojobori T*. (2016) Two-Step Evolution of Endosymbiosis between Hydra and Algae. Mol. Phyl. Evol. 103:19-25.
- Ishikawa M, Yuyama I, Shimizu H, Nozawa M, Ikeo K, Gojobori T*. (2016) Different endosymbiotic interactions inof two hydra species reflect the evolutionary history of endosymbiosis. Genome Biol. Evol. 8:2155-2163.
- Nozawa M*, Fujimi M, Iwamoto C, Onizuka K, Fukuda N, Ikeo K, Gojobori T. (2016) Evolutionary transitions of microRNA-target pairs. Genome Biol. Evol. 8:1621-1633.
- Koga H*, Fujitani H, Morino Y, Miyamoto N, Tsuchimoto J, Shibata TF, Nozawa M, Shigenobu S, Ogura A, Tachibana K, Kiyomoto M, Amemiya S, Wada H. (2016) Experimental approach reveals the role of alx1 in the evolution of the echinoderm larval skeleton. PLOS One 11:e0149067.
- Nozawa M*, Kinjo S. (2016) Origin and evolution of non-coding RNAs. Encyclopedia for Evol. Biol. 3:130-135.
- Kojima KK, Seto Y and Fujiwara H (2016) The Wide Distribution and Change of Target Specificity of R2 Non-LTR Retrotransposons in Animals. PLoS ONE 11:e0163496, 2016
- Satomura K, Tamura K. (2016) Ancient male recombination shaped genetic diversity of neo-Y chromosome in Drosophila albomicans. Mol. Biol. Evol. 33:367-374.
- Liu L, Tamura K, Sanderford M, Gray VE, Kumar S. A (2016) Molecular Evolutionary Reference for the Human Variome. Mol. Biol. Evol. 33:245-254.
- Ohta S, Seto Y, Tamura K, Ishikawa Y, Matsuo T. (2015) Comprehensive identification of odorant-binding protein genes in the seed fly, Delia platura (Diptera: Anthomyiidae). Applied Entomology and Zoology 50:457-463.
- Filipski A, Tamura K, Billing-Ross P, Murillo O, Kumar S. (2015) Phylogenetic placement of metagenomic reads using the minimum evolution principle. BMC Genomics 16:1-9.
- Miyagi R, Akiyama N, Osada N and Takahashi A*. (2015) Complex patterns of cis-regulatory polymorphisms in ebony underlie standing pigmentation variation in Drosophila melanogaster. Mol. Ecol. 24: 5829-5841.
- Tanaka KM, Takahashi A, Fuse N, and Takano-Shimizu-Kouno T. (2014) A novel cell death gene acts to repair patterning defects in Drosophila melanogaster. Genetics 197: 739-742.
- Filipski A, Murillo O, Freydenzon A, Tamura K, Kumar S. (2014) Prospects for building large timetrees using molecular data with incomplete gene coverage among species. Mol. Biol. Evol. 31:2542-2550.
- Takezaki N, Nei M, Tamura K. (2014) POPTREEW: Web version of POPTREE for constructing population trees from allele frequency data and computing some other quantities. Mol. Biol. Evol. 31:1622-1624.
- Stecher G, Liu L, Sanderford M, Peterson P, Tamura K, Kumar S. (2014) MEGA-MD: Molecular Evolutionary Genetics Analysis software with mutational diagnosis of amino acid variation. Bioinformatics 30:1305-1307.
- Ohta S, Seto Y, Tamura K, Ishikawa Y, Matsuo T. (2014) Identification of odorant-binding protein genes expressed in the antennae and the legs of the onion fly, Delia antiqua (Diptera: Anthomyiidae). Appl. Entomol. Zool. 49:89-95.
- Isobe K, Takahashi A, Tamura K. (2013) Cold tolerance and metabolic rate increased by cold acclimation in Drosophila albomicans from natural populations. Genes Genet. Syst. 88:289-300.
- Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. (2013) MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol Biol. Evol. 30:2725-2729.
- Seto Y, Tamura K. (2013) Extensive Differences in Antifungal Immune Response in Two Drosophila Species Revealed by Comparative Transcriptome Analysis. Int. J. Genomics 2013:Article ID 542139.
- Takahashi A. (2013) Pigmentation and behavior: potential association through pleiotropic genes in Drosophila. Genes Genet. Syst. 88: 165-174.
- Tamura K, Battistuzzi FU, Billing-Ross P, Kumar S. (2012) Estimating Divergence Times in Large Molecular Phylogenies. Proc. Nat. Acad. Sci. USA 109:19333-19338.
- Kumar S, Stecher G, Peterson D, Tamura K. (2012) MEGA-CC: Computing Core of Molecular Evolutionary Genetics Analysis program for automated and iterative data analysis. Bioinformatics 28: 2685-2686. 2012.
- Kumar S, Filipski AJ, Battistuzzi FU, Kosakovsky Pond SL, Tamura K. (2012) Statistics and Truth in Phylogenomics. Mol Biol. Evol. 29:457-472.
- Nishimura A, Ishida Y, Takahashi A, Okamoto H, Sakabe M, Itoh M, Takano-Shimizu T, Ozaki M. (2012) Starvation-induced elevation of taste responsiveness and expression of a sugar taste receptor gene in Drosophila melanogaster. J. Neurogenet. 26: 206-15.
- Takahashi A, Fujiwara-Tsujii N, Yamaoka R, Itoh M, Ozaki M, Takano-Shimizu T. (2012) Cuticular hydrocarbon content that affects male mate preference of Drosophila melanogaster from West Africa. Int. J. Evol. Biol. 2012:278903.
|
|
|